Wine Tasting Data
winetaste.RdPaired comparison judgments for two wine tasting studies:
ambilight includes the results of a study on the effect of ambient
lighting on the flavor of wine; redwines includes judgments on the
sensory quality of red wines.
data("winetaste")Format
ambilight A data frame containing 230 observations on 10 variables:
- preference, fruitiness, spiciness, sweetness
Paired comparison of class
paircomp; judgments for one of the 6 ordered pairs of the blue, red, and white lighting conditions.- age
subject age
- gender
factor, subject gender
- sensesmell
self-rating of sense of smell and taste.
- likewine
self-rating of general liking of wine.
- drinkwine
factor, frequency of wine consumption.
- redwhite
factor, preference for red or white wine.
redwines A data frame containing 11 observations on 7 variables:
- bitterness, fruitiness, sourness, roundness, preference
Paired comparison of class
paircomp; judgments for all 10 pairs from 5 red wines: Primitivo di Manduria, Cotes du Rhone, Bourgogne, Shiraz cuvee, Cabernet Sauvignon.- best
factor, Which of the five wines did you like best?
- worst
factor, Which of the five wines did you like worst?
Details
The ambilight data are from Experiment 3 in Oberfeld et al. (2009).
The redwines data were collected among the members of the Sound
Quality Research Unit (SQRU), Department of Acoustics, Aalborg University,
Denmark, in 2004. Details of the red wines are available as an attribute of
the preference variable (see Examples).
References
Oberfeld, D., Hecht, H., Allendorf, U., & Wickelmaier, F. (2009). Ambient lighting modifies the flavor of wine. Journal of Sensory Studies, 24(6), 797–832. doi:10.1111/j.1745-459X.2009.00239.x
Examples
requireNamespace("psychotools")
data(winetaste)
## No effect of ambient lighting on flavor (Oberfeld et al., 2009)
m <- lapply(ambilight[, c("preference", "fruitiness",
"spiciness", "sweetness")],
function(x) eba.order(summary(x, pcmatrix = TRUE)))
lapply(m, summary)
#> $preference
#>
#> Parameter estimates (H0: parameter = 0):
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.32165 0.03731 8.622 <2e-16 ***
#> 2 0.37144 0.04022 9.235 <2e-16 ***
#> 3 0.34565 0.04136 8.356 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Order effects (H0: parameter = 1):
#> Estimate Std. Error z value Pr(>|z|)
#> order 0.50233 0.07032 -7.077 1.47e-12 ***
#> order0 0.50327 0.13972 -3.555 0.000378 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> EBA.order 3 6 -12.623 -11.939 1.370 0.713
#> Order 2 3 -25.451 -12.623 25.654 4.08e-07 ***
#> Effect 1 3 -12.911 -12.623 0.575 0.750
#> Imbalance 1 6 -18.139 -16.444 3.390 0.640
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 31.247
#> Pearson X2: 1.362
#>
#> $fruitiness
#>
#> Parameter estimates (H0: parameter = 0):
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.33887 0.04165 8.137 4.05e-16 ***
#> 2 0.48432 0.04968 9.749 < 2e-16 ***
#> 3 0.36632 0.04668 7.847 4.24e-15 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Order effects (H0: parameter = 1):
#> Estimate Std. Error z value Pr(>|z|)
#> order 0.44932 0.06475 -8.505 < 2e-16 ***
#> order0 0.45570 0.14219 -3.828 0.000129 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> EBA.order 3 6 -14.233 -11.716 5.035 0.169
#> Order 2 3 -30.969 -14.233 33.471 7.23e-09 ***
#> Effect 1 3 -16.071 -14.233 3.675 0.159
#> Imbalance 1 6 -18.139 -16.444 3.390 0.640
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 34.467
#> Pearson X2: 5.01
#>
#> $spiciness
#>
#> Parameter estimates (H0: parameter = 0):
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.37345 0.03725 10.025 <2e-16 ***
#> 2 0.31304 0.03409 9.183 <2e-16 ***
#> 3 0.31375 0.03651 8.593 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Order effects (H0: parameter = 1):
#> Estimate Std. Error z value Pr(>|z|)
#> order 0.9827 0.1299 -0.133 0.894
#> order0 0.9828 0.1319 -0.131 0.896
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> EBA.order 3 6 -12.387 -12.293 0.187 0.980
#> Order 2 3 -12.395 -12.387 0.017 0.895
#> Effect 1 3 -13.000 -12.387 1.227 0.542
#> Imbalance 1 6 -18.139 -16.444 3.390 0.640
#>
#> AIC: 30.773
#> Pearson X2: 0.1872
#>
#> $sweetness
#>
#> Parameter estimates (H0: parameter = 0):
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.30562 0.03492 8.753 < 2e-16 ***
#> 2 0.43390 0.04120 10.532 < 2e-16 ***
#> 3 0.29558 0.03676 8.041 8.95e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Order effects (H0: parameter = 1):
#> Estimate Std. Error z value Pr(>|z|)
#> order 0.69802 0.09466 -3.190 0.00142 **
#> order0 0.70370 0.13392 -2.213 0.02693 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> EBA.order 3 6 -12.845 -12.140 1.411 0.7030
#> Order 2 3 -16.420 -12.845 7.149 0.0075 **
#> Effect 1 3 -15.401 -12.845 5.111 0.0777 .
#> Imbalance 1 6 -18.139 -16.444 3.390 0.6402
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 31.691
#> Pearson X2: 1.411
#>
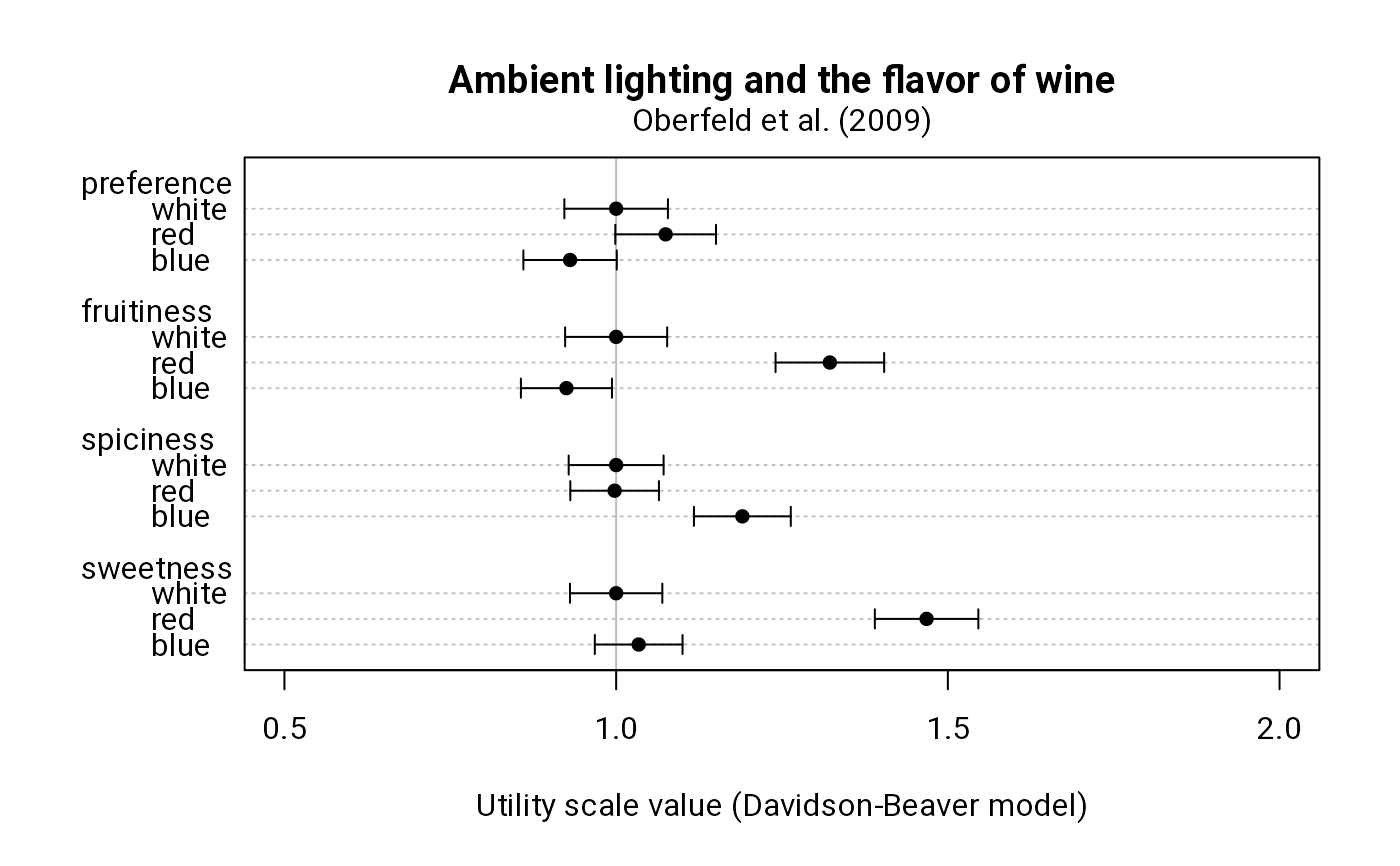
u <- sapply(m, uscale, norm = 3)
dotchart(
u, xlim = c(0.5, 2), pch = 16, panel.first = abline(v = 1, col = "gray"),
main = "Ambient lighting and the flavor of wine",
xlab = "Utility scale value (Davidson-Beaver model)"
)
ci <- sapply(m, function(x) 1.96 * sqrt(diag(cov.u(x))))
arrows(
u - ci, c(16:18, 11:13, 6:8, 1:3), u + ci, c(16:18, 11:13, 6:8, 1:3),
.05, 90, 3)
mtext("Oberfeld et al. (2009)", line = 0.5)
 ## Sensory quality of red wines
psychotools::covariates(redwines$preference) # details of the wines
#> code grape country name
#> Primitivo A Primitivo di Manduria Italy Faunus Vendemmia
#> CotesDuRhone B Cotes du Rhone France <NA>
#> Bourgogne C Bourgogne France <NA>
#> ShirazCuvee D Shiraz cuvee South Africa Cape Red
#> CabernetSauvignon E Cabernet Sauvignon Chile Santiago 1541
#> winery year alc.prc price.dkk
#> Primitivo <NA> 2001 14.0 53
#> CotesDuRhone Domaine des Estremieres 2001 14.0 55
#> Bourgogne Domaine Bernard Moreau et Fils 2002 12.5 110
#> ShirazCuvee Drostdy Hof 2003 13.5 33
#> CabernetSauvignon D. O. Colchagua Valley 2003 12.5 45
m <- lapply(redwines[, c("bitterness", "fruitiness", "sourness",
"roundness", "preference")],
function(x) eba(summary(x, pcmatrix = TRUE)))
lapply(m, summary)
#> $bitterness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.21271 0.05094 4.176 2.97e-05 ***
#> 2 0.16895 0.04288 3.940 8.15e-05 ***
#> 3 0.26972 0.05985 4.507 6.58e-06 ***
#> 4 0.12447 0.03391 3.671 0.000242 ***
#> 5 0.07688 0.02364 3.252 0.001144 **
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -43.02 -34.87 16.31 0.6367
#> EBA 4 10 -15.89 -13.61 4.55 0.6027
#> Effect 0 4 -21.77 -15.89 11.76 0.0193 *
#> Imbalance 1 10 -21.25 -21.25 0.00 1.0000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 39.775
#> Pearson X2: 4.35
#>
#> $fruitiness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.13684 0.03676 3.722 0.000197 ***
#> 2 0.15921 0.04129 3.856 0.000115 ***
#> 3 0.10019 0.02905 3.449 0.000563 ***
#> 4 0.25113 0.05801 4.329 1.50e-05 ***
#> 5 0.27159 0.06117 4.440 8.99e-06 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -41.658 -35.124 13.068 0.8351
#> EBA 4 10 -15.930 -13.870 4.121 0.6604
#> Effect 0 4 -20.404 -15.930 8.947 0.0624 .
#> Imbalance 1 10 -21.255 -21.255 0.000 1.0000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 39.86
#> Pearson X2: 3.937
#>
#> $sourness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.11183 0.03399 3.290 0.001002 **
#> 2 0.15359 0.04400 3.491 0.000482 ***
#> 3 0.40275 0.07768 5.185 2.16e-07 ***
#> 4 0.08796 0.02797 3.145 0.001664 **
#> 5 0.06860 0.02295 2.989 0.002795 **
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -46.109 -34.555 23.108 0.232608
#> EBA 4 10 -14.478 -13.301 2.354 0.884431
#> Effect 0 4 -24.855 -14.478 20.754 0.000354 ***
#> Imbalance 1 10 -21.255 -21.255 0.000 1.000000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 36.955
#> Pearson X2: 2.368
#>
#> $roundness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.19038 0.04799 3.967 7.28e-05 ***
#> 2 0.12822 0.03553 3.609 0.000308 ***
#> 3 0.04901 0.01758 2.788 0.005305 **
#> 4 0.26312 0.06016 4.374 1.22e-05 ***
#> 5 0.22341 0.05390 4.145 3.41e-05 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -46.802 -34.484 24.638 0.172810
#> EBA 4 10 -15.577 -13.229 4.695 0.583437
#> Effect 0 4 -25.548 -15.577 19.943 0.000513 ***
#> Imbalance 1 10 -21.255 -21.255 0.000 1.000000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 39.153
#> Pearson X2: 4.684
#>
#> $preference
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.14987 0.03946 3.798 0.000146 ***
#> 2 0.12857 0.03503 3.671 0.000242 ***
#> 3 0.07342 0.02289 3.208 0.001336 **
#> 4 0.30590 0.06469 4.728 2.26e-06 ***
#> 5 0.17469 0.04441 3.934 8.37e-05 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -44.431 -34.838 19.187 0.4449
#> EBA 4 10 -16.732 -13.583 6.297 0.3908
#> Effect 0 4 -23.176 -16.732 12.889 0.0118 *
#> Imbalance 1 10 -21.255 -21.255 0.000 1.0000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 41.463
#> Pearson X2: 6.376
#>
u <- sapply(m, uscale)
dotchart(
u[order(u[, "preference"]), ], log = "x",
panel.first = abline(v = 1/5, col = "gray"),
main = "SQRU red wine tasting",
xlab = "Utility scale value (BTL model), choice proportion (+)"
)
points(as.vector(
prop.table(table(redwines$best))[order(u[, "preference"])]
), 1:5, pch = 3)
## Sensory quality of red wines
psychotools::covariates(redwines$preference) # details of the wines
#> code grape country name
#> Primitivo A Primitivo di Manduria Italy Faunus Vendemmia
#> CotesDuRhone B Cotes du Rhone France <NA>
#> Bourgogne C Bourgogne France <NA>
#> ShirazCuvee D Shiraz cuvee South Africa Cape Red
#> CabernetSauvignon E Cabernet Sauvignon Chile Santiago 1541
#> winery year alc.prc price.dkk
#> Primitivo <NA> 2001 14.0 53
#> CotesDuRhone Domaine des Estremieres 2001 14.0 55
#> Bourgogne Domaine Bernard Moreau et Fils 2002 12.5 110
#> ShirazCuvee Drostdy Hof 2003 13.5 33
#> CabernetSauvignon D. O. Colchagua Valley 2003 12.5 45
m <- lapply(redwines[, c("bitterness", "fruitiness", "sourness",
"roundness", "preference")],
function(x) eba(summary(x, pcmatrix = TRUE)))
lapply(m, summary)
#> $bitterness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.21271 0.05094 4.176 2.97e-05 ***
#> 2 0.16895 0.04288 3.940 8.15e-05 ***
#> 3 0.26972 0.05985 4.507 6.58e-06 ***
#> 4 0.12447 0.03391 3.671 0.000242 ***
#> 5 0.07688 0.02364 3.252 0.001144 **
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -43.02 -34.87 16.31 0.6367
#> EBA 4 10 -15.89 -13.61 4.55 0.6027
#> Effect 0 4 -21.77 -15.89 11.76 0.0193 *
#> Imbalance 1 10 -21.25 -21.25 0.00 1.0000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 39.775
#> Pearson X2: 4.35
#>
#> $fruitiness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.13684 0.03676 3.722 0.000197 ***
#> 2 0.15921 0.04129 3.856 0.000115 ***
#> 3 0.10019 0.02905 3.449 0.000563 ***
#> 4 0.25113 0.05801 4.329 1.50e-05 ***
#> 5 0.27159 0.06117 4.440 8.99e-06 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -41.658 -35.124 13.068 0.8351
#> EBA 4 10 -15.930 -13.870 4.121 0.6604
#> Effect 0 4 -20.404 -15.930 8.947 0.0624 .
#> Imbalance 1 10 -21.255 -21.255 0.000 1.0000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 39.86
#> Pearson X2: 3.937
#>
#> $sourness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.11183 0.03399 3.290 0.001002 **
#> 2 0.15359 0.04400 3.491 0.000482 ***
#> 3 0.40275 0.07768 5.185 2.16e-07 ***
#> 4 0.08796 0.02797 3.145 0.001664 **
#> 5 0.06860 0.02295 2.989 0.002795 **
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -46.109 -34.555 23.108 0.232608
#> EBA 4 10 -14.478 -13.301 2.354 0.884431
#> Effect 0 4 -24.855 -14.478 20.754 0.000354 ***
#> Imbalance 1 10 -21.255 -21.255 0.000 1.000000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 36.955
#> Pearson X2: 2.368
#>
#> $roundness
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.19038 0.04799 3.967 7.28e-05 ***
#> 2 0.12822 0.03553 3.609 0.000308 ***
#> 3 0.04901 0.01758 2.788 0.005305 **
#> 4 0.26312 0.06016 4.374 1.22e-05 ***
#> 5 0.22341 0.05390 4.145 3.41e-05 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -46.802 -34.484 24.638 0.172810
#> EBA 4 10 -15.577 -13.229 4.695 0.583437
#> Effect 0 4 -25.548 -15.577 19.943 0.000513 ***
#> Imbalance 1 10 -21.255 -21.255 0.000 1.000000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 39.153
#> Pearson X2: 4.684
#>
#> $preference
#>
#> Parameter estimates:
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.14987 0.03946 3.798 0.000146 ***
#> 2 0.12857 0.03503 3.671 0.000242 ***
#> 3 0.07342 0.02289 3.208 0.001336 **
#> 4 0.30590 0.06469 4.728 2.26e-06 ***
#> 5 0.17469 0.04441 3.934 8.37e-05 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> Overall 1 20 -44.431 -34.838 19.187 0.4449
#> EBA 4 10 -16.732 -13.583 6.297 0.3908
#> Effect 0 4 -23.176 -16.732 12.889 0.0118 *
#> Imbalance 1 10 -21.255 -21.255 0.000 1.0000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 41.463
#> Pearson X2: 6.376
#>
u <- sapply(m, uscale)
dotchart(
u[order(u[, "preference"]), ], log = "x",
panel.first = abline(v = 1/5, col = "gray"),
main = "SQRU red wine tasting",
xlab = "Utility scale value (BTL model), choice proportion (+)"
)
points(as.vector(
prop.table(table(redwines$best))[order(u[, "preference"])]
), 1:5, pch = 3)
