Elimination-by-Aspects (EBA) Models with Order-Effect
eba.order.RdFits a (multi-attribute) probabilistic choice model that accounts for the effect of the presentation order within a pair.
Arguments
- M1, M2
two square matrices or data frames consisting of absolute choice frequencies in both within-pair orders; row stimuli are chosen over column stimuli. If M2 is empty (default), M1 is assumed to be a 3d array containing both orders
- A
see
eba- s
the starting vector with default
1/Jfor allJaspect parameters, and1for the order effect- constrained
see
eba- object
an object of class
eba.order, typically the result of a call toeba.order- ...
additional arguments
Details
The choice models include a single multiplicative order effect,
order, that is constant for all pairs (see Davidson and Beaver,
1977). An order effect < 1 (> 1) indicates a bias in favor of the first
(second) interval. See eba for choice models without order
effect.
Several likelihood ratio tests are performed (see also
summary.eba).
EBA.order tests an order-effect EBA model against a saturated
binomial model; this corresponds to a goodness of fit test of the former
model.
Order tests an EBA model with an order effect constrained to 1
against an unconstrained order-effect EBA model; this corresponds to a test
of the order effect.
Effect tests an order-effect indifference model (where all scale
values are equal, but the order effect is free) against the order-effect EBA
model; this corresponds to testing for a stimulus effect; order0 is
the estimate of the former model.
Wickelmaier and Choisel (2006) describe a model that generalizes the Davidson-Beaver model and allows for an order effect in Pretree and EBA models.
Value
- coefficients
a vector of parameter estimates, the last component holds the order-effect estimate
- estimate
same as
coefficients- logL.eba
the log-likelihood of the fitted model
- logL.sat
the log-likelihood of the saturated (binomial) model
- goodness.of.fit
the goodness of fit statistic including the likelihood ratio fitted vs. saturated model (-2logL), the degrees of freedom, and the p-value of the corresponding chi-square distribution
- u.scale
the unnormalized utility scale of the stimuli; each utility scale value is defined as the sum of aspect values (parameters) that characterize a given stimulus
- hessian
the Hessian matrix of the likelihood function
- cov.p
the covariance matrix of the model parameters
- chi.alt
the Pearson chi-square goodness of fit statistic
- fitted
3d array of the fitted paired-comparison matrices
- y1
the data vector of the upper triangle matrices
- y0
the data vector of the lower triangle matrices
- n
the number of observations per pair (
y1 + y0)- mu
the predicted choice probabilities for the upper triangles
- M1, M2
the data matrices
References
Davidson, R.R., & Beaver, R.J. (1977). On extending the Bradley-Terry model to incorporate within-pair order effects. Biometrics, 33, 693–702.
Wickelmaier, F., & Choisel, S. (2006). Modeling within-pair order effects in paired-comparison judgments. In D.E. Kornbrot, R.M. Msetfi, & A.W. MacRae (eds.), Fechner Day 2006. Proceedings of the 22nd Annual Meeting of the International Society for Psychophysics (p. 89–94). St. Albans, UK: The ISP.
See also
Examples
data(heaviness) # weights judging data
ebao1 <- eba.order(heaviness) # Davidson-Beaver model
summary(ebao1) # goodness of fit
#>
#> Parameter estimates (H0: parameter = 0):
#> Estimate Std. Error z value Pr(>|z|)
#> 1 0.013623 0.002390 5.700 1.20e-08 ***
#> 2 0.028221 0.004352 6.484 8.95e-11 ***
#> 3 0.065548 0.008816 7.435 1.04e-13 ***
#> 4 0.185365 0.020133 9.207 < 2e-16 ***
#> 5 0.397046 0.025805 15.386 < 2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Order effects (H0: parameter = 1):
#> Estimate Std. Error z value Pr(>|z|)
#> order 1.33734 0.11553 2.920 0.00350 **
#> order0 1.17391 0.06345 2.741 0.00613 **
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Model tests:
#> Df1 Df2 logLik1 logLik2 Deviance Pr(>Chi)
#> EBA.order 5 20 -39.250 -35.966 6.567 0.968574
#> Order 4 5 -45.025 -39.250 11.550 0.000677 ***
#> Effect 1 5 -296.290 -39.250 514.081 < 2e-16 ***
#> Imbalance 1 20 -57.532 -57.532 0.000 1.000000
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> AIC: 88.5
#> Pearson X2: 6.602
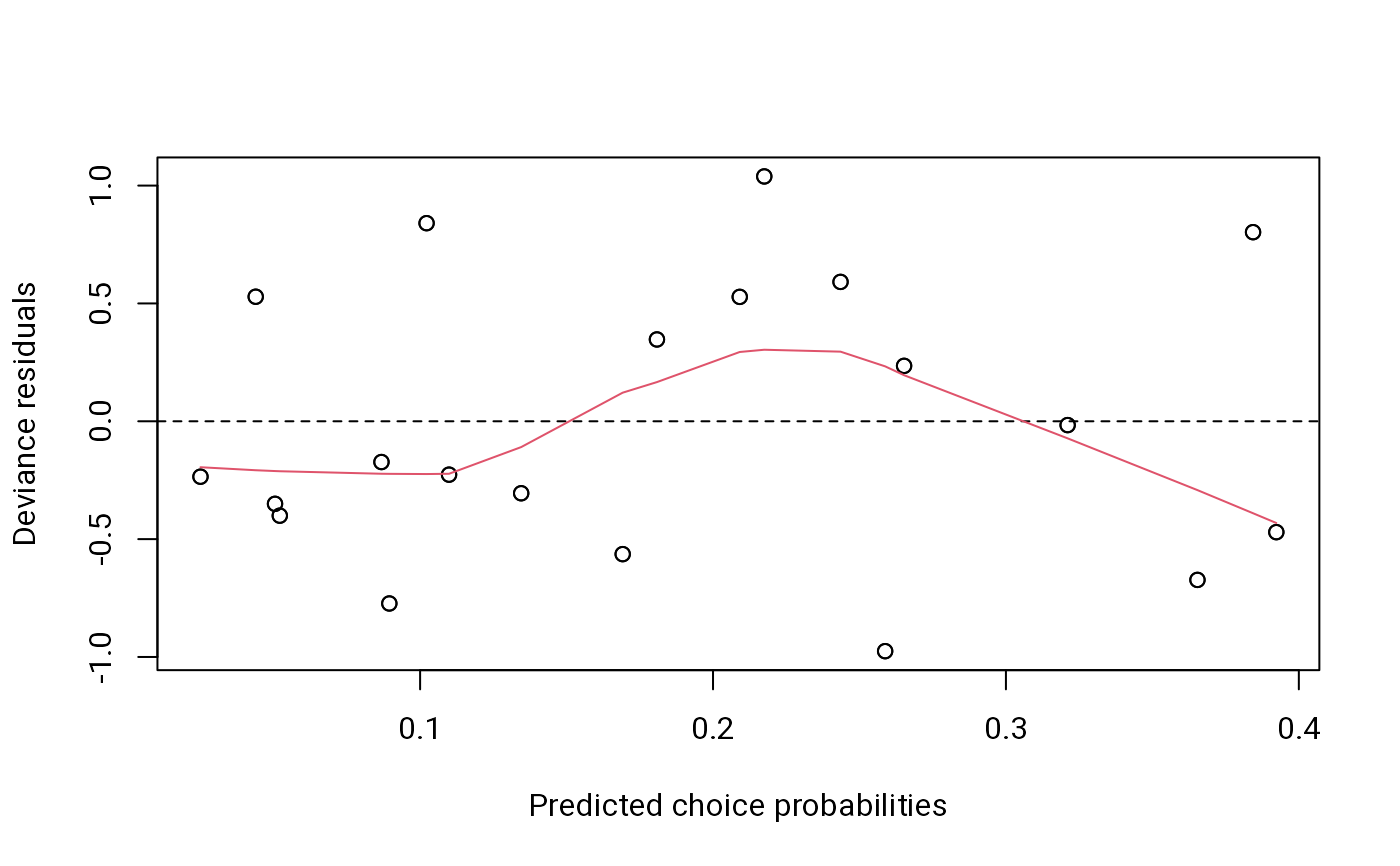
plot(ebao1) # residuals versus predicted values
 confint(ebao1) # confidence intervals for parameters
#> 2.5 % 97.5 %
#> 1 0.008939213 0.01830734
#> 2 0.019689912 0.03675139
#> 3 0.048269446 0.08282664
#> 4 0.145905589 0.22482461
#> 5 0.346468731 0.44762381
#> order 1.110902126 1.56378025
confint(ebao1) # confidence intervals for parameters
#> 2.5 % 97.5 %
#> 1 0.008939213 0.01830734
#> 2 0.019689912 0.03675139
#> 3 0.048269446 0.08282664
#> 4 0.145905589 0.22482461
#> 5 0.346468731 0.44762381
#> order 1.110902126 1.56378025