Diagnostic Plot for EBA Models
plot.eba.RdPlots elimination-by-aspects (EBA) model residuals against fitted values.
# S3 method for class 'eba'
plot(x, xlab = "Predicted choice probabilities",
ylab = "Deviance residuals", ...)Arguments
- x
an object of class
eba, typically the result of a call toeba- xlab, ylab, ...
graphical parameters passed to plot.
Details
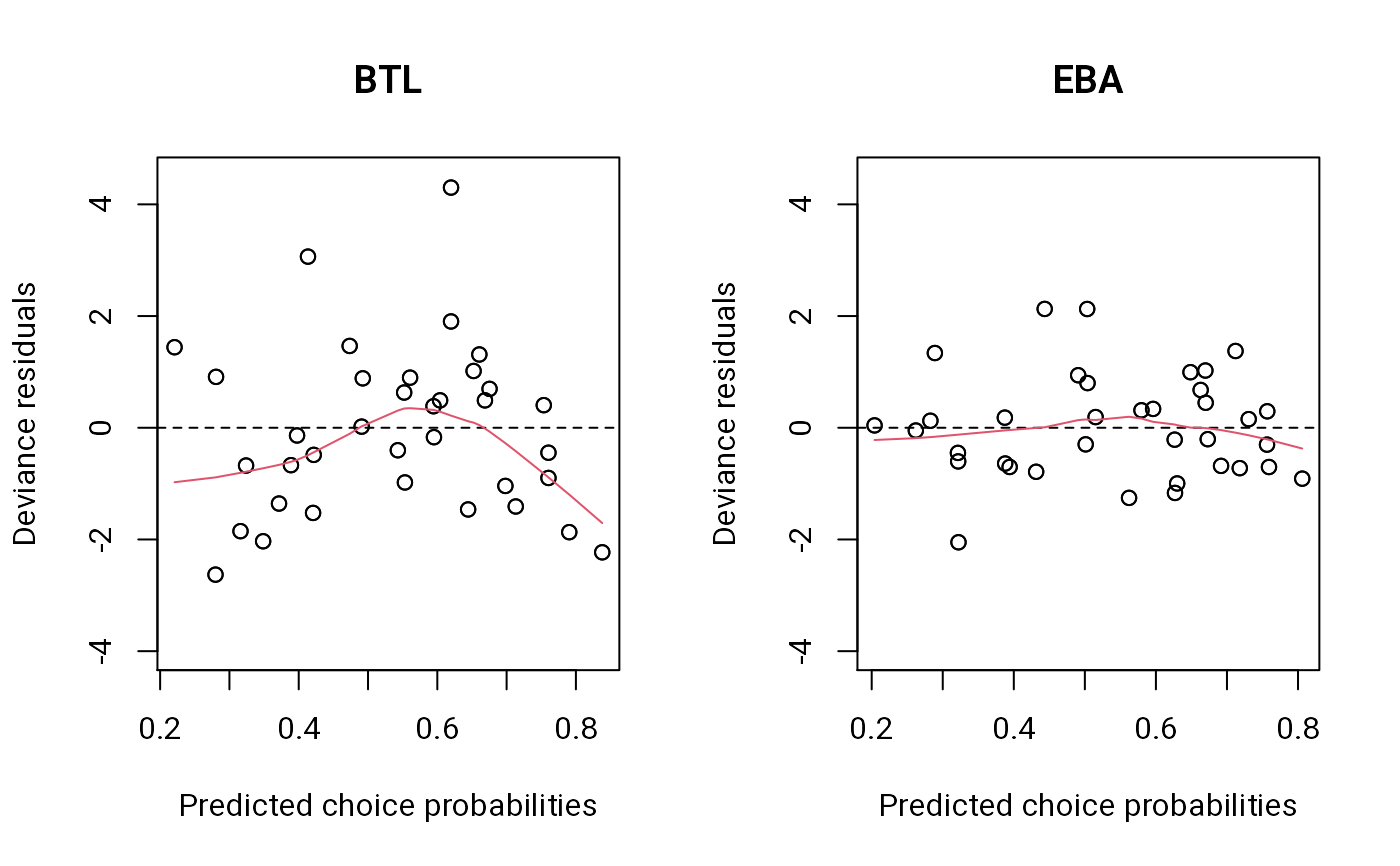
The deviance residuals are plotted against the predicted choice probabilities for the upper triangle of the paired-comparison matrix.
See also
Examples
## Compare two choice models
data(celebrities) # absolute choice frequencies
btl1 <- eba(celebrities) # fit Bradley-Terry-Luce model
A <- list(c(1,10), c(2,10), c(3,10),
c(4,11), c(5,11), c(6,11),
c(7,12), c(8,12), c(9,12)) # the structure of aspects
eba1 <- eba(celebrities, A) # fit elimination-by-aspects model
anova(btl1, eba1) # model comparison based on likelihoods
#> Analysis of Deviance Table
#>
#> Model 1: btl1
#> Model 2: eba1
#> Resid. Df Resid. Dev Df Deviance Pr(>Chi)
#> 1 28 78.217
#> 2 25 30.166 3 48.051 2.077e-10 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
par(mfrow = 1:2) # residuals versus fitted values
plot(btl1, main = "BTL", ylim = c(-4, 4.5)) # BTL doesn't fit well
plot(eba1, main = "EBA", ylim = c(-4, 4.5)) # EBA fits better