Diagnostic Plot for MPT Models
plot.mpt.RdPlots MPT residuals against fitted values.
Arguments
- x, object
an object of class
mpt, typically the result of a call tompt.- showNames
logical. Should the names of the residuals be plotted? Defaults to
TRUE.- xlab, ylab
graphical parameters passed to plot.
- type
the type of residuals which should be returned; the alternatives are:
"deviance"(default) and"pearson".- ...
further arguments passed to or from other methods.
Details
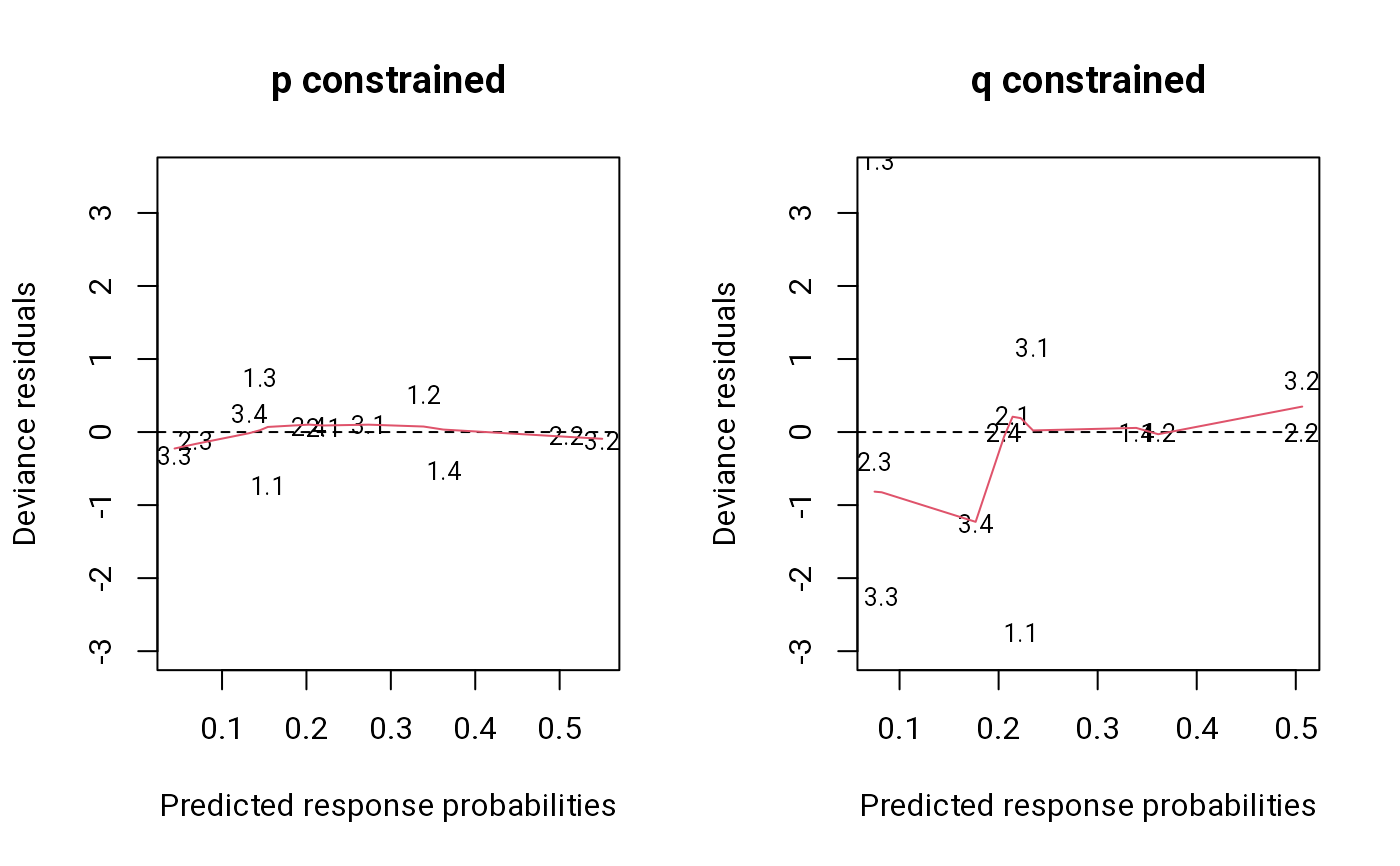
The deviance residuals are plotted against the predicted response
probabilities. If showNames is true, plotting symbols are the
names of the residuals.
Value
For residuals, a named vector of residuals having as many elements as
response categories.
See also
Examples
## Compare two constrained MPT models
data(proact)
spec <- mptspec(
p1*q1*r1,
p1*q1*(1 - r1),
p1*(1 - q1)*r1,
(1 - p1) + p1*(1 - q1)*(1 - r1),
p2*q2*r2,
p2*q2*(1 - r2),
p2*(1 - q2)*r2,
(1 - p2) + p2*(1 - q2)*(1 - r2),
p3*q3*r3,
p3*q3*(1 - r3),
p3*(1 - q3)*r3,
(1 - p3) + p3*(1 - q3)*(1 - r3)
)
m1 <- mpt(update(spec, .restr = list(p2=p1, p3=p1)),
proact[proact$test == 1, ])
m2 <- mpt(update(spec, .restr = list(q2=q1, q3=q1)), m1$y)
par(mfrow = c(1, 2)) # residuals versus fitted values
plot(m1, main = "p constrained", ylim = c(-3, 3.5)) # good fit
plot(m2, main = "q constrained", ylim = c(-3, 3.5)) # bad fit
 sum( resid(m1)^2 ) # likelihood ratio G2
#> [1] 1.845292
sum( resid(m1, "pearson")^2 ) # Pearson X2
#> [1] 1.845037
sum( resid(m1)^2 ) # likelihood ratio G2
#> [1] 1.845292
sum( resid(m1, "pearson")^2 ) # Pearson X2
#> [1] 1.845037