Moral Dilemma Judgment
moraldilemma.RdHennig and Huetter (2020) proposed a multinomial model of moral dilemma judgment and evaluated the model in a series of experiments. Participants were presented with hypothetical scenarios that required a decision whether or not to break a moral norm.
Berentelg (2020) conducted a replication study that was designed to be similar to Experiment 2b in Hennig and Huetter (2020).
data(moraldilemma)Format
MDHennig2020 A data frame consisting of seven variables:
selfrelfactor. Instructions about self-relevant consequences of the decision where either (
absent) or (present).congrcyfactor. Endorsement of consequences and norm endorsement lead to different decisions (
incongruent) or to the same decision (congruent).defaultfactor. The norm may be adhered to by continuing (
inactiondefault state) or by changing (actiondefault state) an ongoing behavior.breaknormfactor. Decision to break the norm.
exp1,exp2bthe aggregate response frequencies for Experiment 1 and 2b, respectively.
treeidan identifier for the single trees of the joint multinomial model.
MDreplication A data frame containing 751 observations of five
variables:
selfrelfactor. See above.
genderfactor. Participant gender.
ageparticipant age.
rtmedian response time (in seconds) across scenarios.
ya matrix of response frequencies per participant. Each column represents a combination of the factors
congrcy,default, andbreaknorm.
Source
Hennig, M., & Huetter, M. (2020). Revisiting the divide between deontology and utilitarianism in moral dilemma judgment: A multinomial modeling approach. Journal of Personality and Social Psychology, 118(1), 22–56. doi:10.1037/pspa0000173
Berentelg, M. (2020). Multinomial modeling of moral dilemma judgment: A replication study. Bachelor thesis. University of Tuebingen, Germany. https://osf.io/mb32t/
See also
mpt.
Examples
data(moraldilemma)
## Exp. 1: proCNI and process dissociation (PD) model
s <- mptspec("proCNI")
exp1 <- subset(MDHennig2020, selfrel == "absent")
mpt(update(s, .restr = list(J=I)), data = exp1, freqvar = "exp1")
#>
#> Multinomial processing tree (MPT) models
#>
#> Parameter estimates:
#> C N I
#> 0.4215 0.5273 0.5601
#>
#> Goodness of fit (2 log likelihood ratio):
#> G2(1) = 1.183, p = 0.2768
#>
mpt(update(s, .restr = list(I=0, J=1)), data = exp1, freqvar = "exp1")
#>
#> Multinomial processing tree (MPT) models
#>
#> Parameter estimates:
#> C N
#> 0.4202 0.7615
#>
#> Goodness of fit (2 log likelihood ratio):
#> G2(2) = 1.814, p = 0.4038
#>
## Exp. 2b: self-relevant consequences and norm endorsement
s <- mptspec("proCNI", .replicates = 2, .restr = list(J1=I1, J2=I2))
m1 <- mpt(s, data = MDHennig2020, freqvar = "exp2b")
m2 <- mpt(update(m1$spec, .restr = list(N1=N2)), data = m1$y)
anova(m2, m1)
#> Analysis of Deviance Table
#>
#> Model 1: m2
#> Model 2: m1
#> Resid. Df Resid. Dev Df Deviance Pr(>Chi)
#> 1 3 8.7267
#> 2 2 0.5721 1 8.1546 0.004295 **
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
## Replication of Exp. 2b
md.agg <- aggregate(y ~ selfrel, MDreplication, sum)
y <- as.vector(t(md.agg[, -1]))
m3 <- mpt(s, data = y)
m4 <- mpt(update(s, .restr = list(N1=N2)), data = y)
anova(m4, m3)
#> Analysis of Deviance Table
#>
#> Model 1: m4
#> Model 2: m3
#> Resid. Df Resid. Dev Df Deviance Pr(>Chi)
#> 1 3 24.018
#> 2 2 21.671 1 2.3473 0.1255
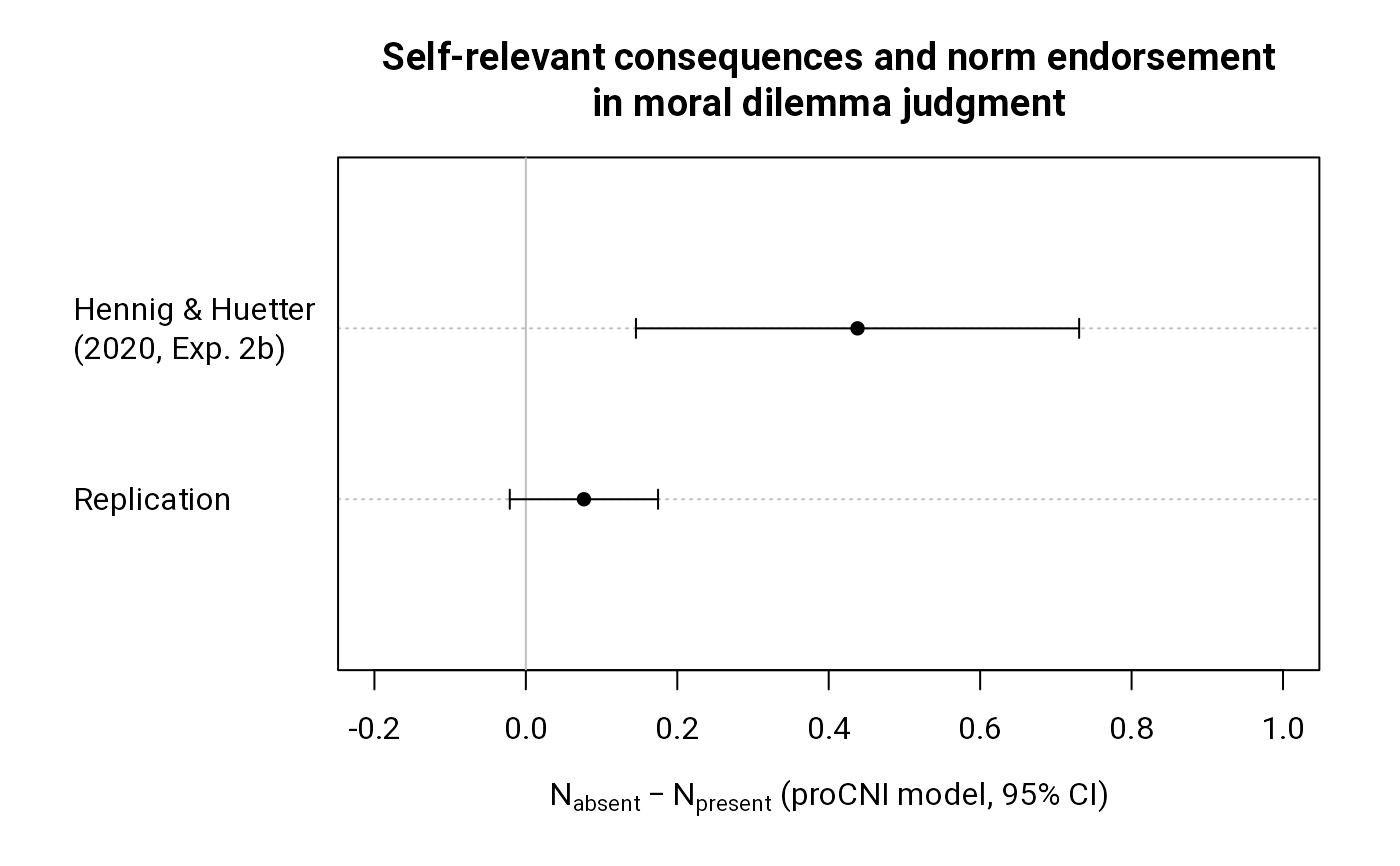
coefs <- c(diff(coef(m3)[c("N2", "N1")]),
diff(coef(m1)[c("N2", "N1")]))
names(coefs) <- c("Replication", "Hennig & Huetter\n(2020, Exp. 2b)")
ci <- coefs + rbind(
qnorm(c(.025, .975))*sqrt(sum(diag(vcov(m3))[c("N2", "N1")])),
qnorm(c(.025, .975))*sqrt(sum(diag(vcov(m1))[c("N2", "N1")]))
)
dotchart(coefs, pch = 16, xlim = c(-.2, 1),
xlab = expression(N[absent] - N[present]~"(proCNI model, 95% CI)"),
main = paste("Self-relevant consequences and norm endorsement",
"in moral dilemma judgment", sep = "\n"))
abline(v = 0, col = "gray")
arrows(ci[, 1], 1:2, ci[, 2], 1:2, .05, 90, 3)